Lab Focus
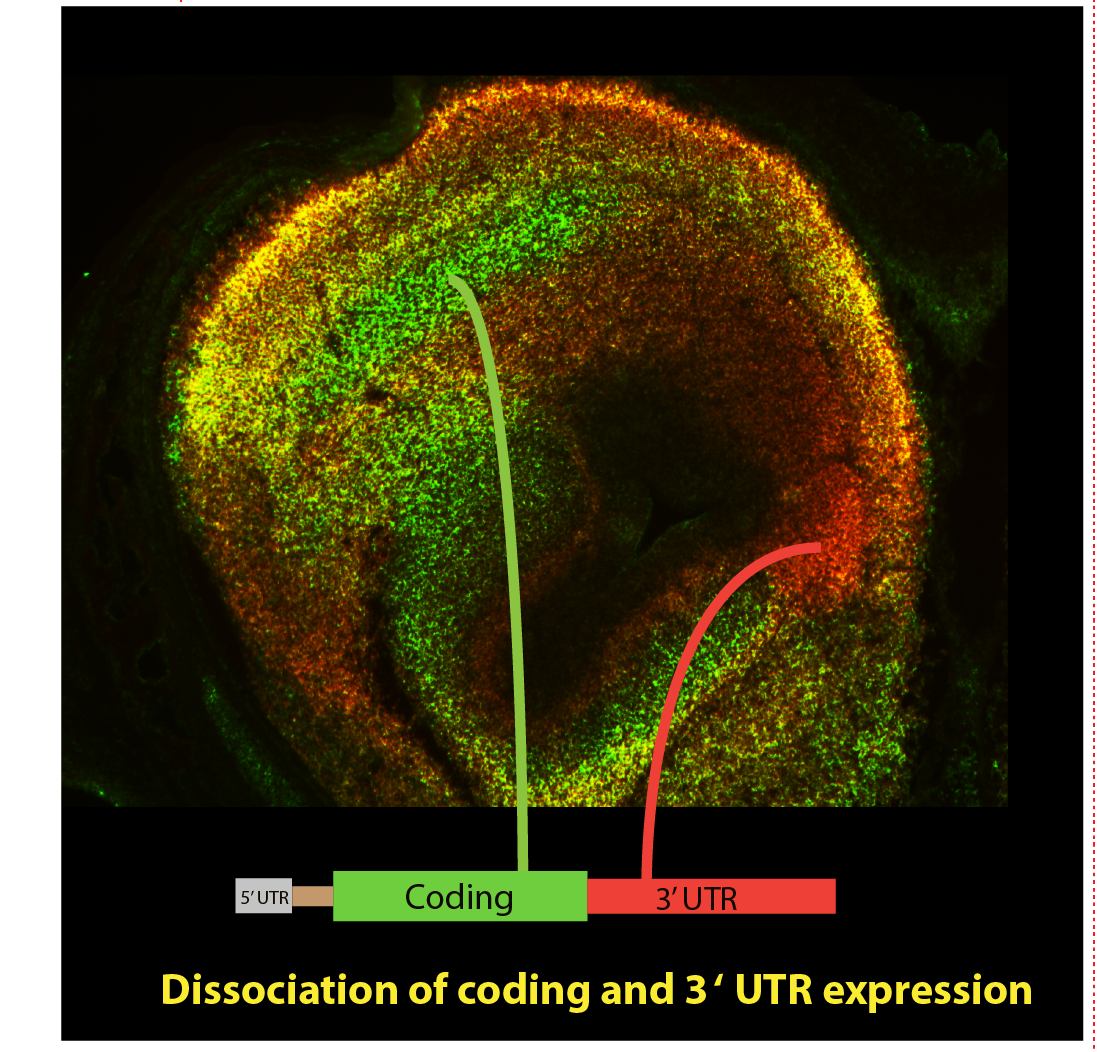
While examining transcriptomes of embryonic dopaminergic neurons we made the surprising discovery of the widespread expression of isolated 3’ UTR sequences (i3'UTRs); 3'UTRs (red signal in above picture), expressed in the absence of their cognate coding sequences (CDS) (green in above picture) and (Kocabas et al, 2015).
Our analyses showed that differential 3'UTR/CDS expression is not particular to development or to DA neurons but occurs in all tissues and at all ages examined.
By two-color in situ hybridization, for every gene examined (>50 to date), we observe differential CDS and cognate 3’ UTR expression in some cells across a panel of tissues.
This differential 3’UTR/CDS expression appears purposeful (not random) in that it is consistent in particular cells across animals, shows exquisite, spatially graded patterns, and changes with developmental or biological state- all suggestive of important functions for these non-coding UTR RNA species.
Further, when cognate 3’UTR and CDS sequences are co-expressed, they may be localized to different subcellular compartments.
Although these findings contradict traditional dogma which suggests that coding regions and UTRs are made as a unit and remain together until degradation- similar results were reported first in 2011 (Mercer et al, 2011), and verified recently in late 2017 (Malka et al, 2017) by independent labs; the former found discordant 3'UTR/CDS expression across human, mouse and drosophila transcriptomes and the latter suggests that thousands of isolated 3'UTR species exist at a given time.
Our initial studies raise a number of questions, most importantly how such isolated 3'UTRs species arise and secondly whether they play important or critical biological roles. Our lab focuses on the biological role of isolated 3'UTRs using three model paradigms (i) neural stem cells (ii) hair follicle/skin stem cells and iii) neural differentiation, in particular dopamine neurons, cortical neurons and the peripheral sensory neurons.
Ji, S., Yang, Z., Gozali, L., Kenney, T., Kocabas, A., Park, C.J., Hynes, M. (2021) Distinct expression of select and transcriptome-wide isolated 3’UTRs suggests critical roles in development and transition states. PLoS One 16 (5).
Kocabas, A., Duarte, T., Kumar, S. & Hynes, M. A. Widespread Differential Expression of Coding Region and 3' UTR Sequences in Neurons and Other Tissues. Neuron 88, 1149-1156, doi:10.1016/j.neuron.2015.10.048 (2015).
Mercer, T. R. et al. Expression of distinct RNAs from 3' untranslated regions. Nucleic Acids Res 39,2393-2403, doi:10.1093/nar/gkq1158 (2011).
Malka, Y. et al. Post-transcriptional 3 -UTR cleavage of mRNA transcripts generates thousands of stable uncapped autonomous RNA fragments. Nat Commun 8, 2029, doi:10.1038/s41467-017-02099-7 (2017).